
|
On the Role of Stop Codons in the Genetic Code |
|
John Cole |

|
Overview: Description & Motivation |
|
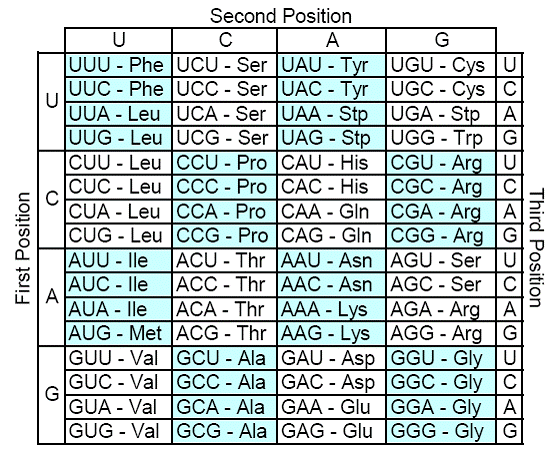
The genetic code is ubiquitous across essentially all life. It is read in triplets of the four standard RNA nucleotides (U, C, G, A), called codons, with each unique codon specifying one of the 20 amino acids to be added in sequence to a protein. There are 64 total codons coding for these 20 amino acids so there is significant degeneracy within the code, meaning multiple codons generally code for the same amino acid. Among the 64 possible codons, three “stop codons” (UAA, UAG, and UGA) do not specify any amino acid, and instead signal the end of a protein. In the case of the wild type code, single nucleotide substitutions (SNS's) often have no effect on a specified amino acid; for instance the codon CUG, specifying leucine, can undergo any substitution in the third base position (CUU, CUC, CUA) as well as a first position substitution to UUG with no effect on the resulting amino acid. The wild type genetic code appears to go further than merely conserving specific amino acids; it appears to conserve physico-chemical properties as well by grouping chemically similar amino acids together in “codon-space”. In the event that a given codon specifying one amino acid undergoes an SNS to become a codon specifying a different amino acid, our genetic code is set up so that the two amino acids are often chemically very similar. The most important chemical property in this respect is an amino acids' polar requirement , a measure of its hydrophobicity or -philicity (Haig 1991).
The question I seek to answer is: if the chemical properties of the amino acids determine their locations in codon-space, what determines the locations of the three stop codons? The focus of this investigation is on determining what possible purpose, if any, the stop codons serve via their locations in codon-space.
|

|
A small pocket organizer, similar in many respects to the netbook I used to run the code developed herein. |
|
The wild type genetic code: |