
|
On the Role of Stop Codons in the Genetic Code |
|
John Cole |

|
A preliminary set of calculations: |
|
†††††††††††††††† As a preliminary step and a proof of concept, the steady state eigenvectors were calculated for 1000 random genetic codes.† In each case the genetic code considered had the same overall codon frequencies (ie. 6 leucine codons, 3 stop codons, etc.) as the wild type code.† These eigenvectors were used to calculate the respective expectation values for the number of heavy atoms, N,† mollecular mass, M, and ratios† Hf / N and† Hf / M.† As well, a measure of the afore mentioned polar requirement conservation,† ΔPódefined as the sum over all amino acids of the absolute values of the changes in polar requirement of a given amino acid converting to another amino acid under a SNSówas also calculated for each random code.† The means and standard deviations of each of these five values were calculated and compared to the values for the wild type code:
†††††††††††††††† We see that the results are encouraging.† As expected, the wild type genetic code does appear incredibly effective in minimizing ΔPówith a value more than 12 sigma better than that of a random code.† Albeit not as striking, all four of the proposed measures of mollecular cost also appear unlikely to occur randomly.† M and N appear to be the strongest in this regard, both of which have expectation values lower than more than an estimated 99% of random codes (assuming a normal distribution).† M appears to be the best optimized by the wild type code of all four measures considered. |
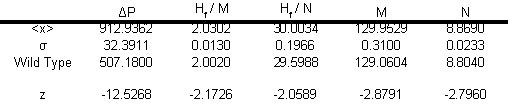
|
Data comparing a set of 1000 random genetic codes with the wild type code with respect to our four molecular cost measures: |