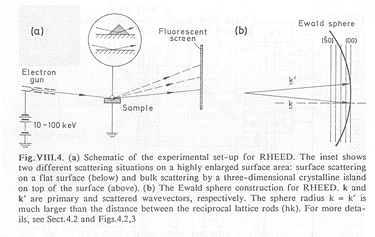
(Fig. 1) RHEED method
SIMULATION OF MOLECULAR BEAM
HOMOEPITAXIAL GROWTH OF GAAS
1. OVERVIEW
1.1 ISLAND FORMATION AND RHEED AMPLITUDE IN MBE
Molecular Beam Epitaxy (MBE) is used to grow monocrystalline thin films. The stoichiometry can mostly be controlled by the growth parameters temperature and partial pressure of the constituents. The RHEED technique (Fig. 1) is used to check the crystallographic structure of the epitaxial surface during the growth process. An excess of one of the constituents will lead to non-integral-order spots between the regular Bragg spots in the diffraction pattern.

(Fig. 1) RHEED method
The intensity of the Bragg spots shows oscillations with a regular period (Fig. 2). The decrease in Bragg intensity is due to an increase of background intensity, which is caused by the surface roughness. The surface roughness corresponds to the number of non-finished layers in the growth process.
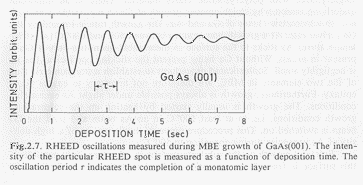
(Fig. 2) RHEED oscillations
I simulated the initial stage of GaAs
homoepitaxial growth, where the island size of adatoms determines
the surface roughness.
1.2 SIMULATION
I used the Metropolis Monte Carlo method with 2-dim periodic boundary conditions for the simulation. The setup was a perfect GaAs crystal of 6 x 6 x 1 unit cells. Two layers at the bottom of the simulation cell were fixed to model a rigid crystal. The molecular beam was modelled by putting a certain amount of Ga and As atoms close to the surface (Fig. 3). The number of adatoms is determined by the coverage ratio (Number of adatoms/Number of atoms in a layer).
![[Graphics:Images/index_gr_1.gif]](initialc.gif)
(Fig. 3) Typical initial configuration.
As a potential for GaAs I took a modified Tersoff potential (1). The modification is an additional Coulomb term as discussed in [p1]. In order to simplify the calculation I used a shielded Coulomb potential, where the parameter k is chosen to make it vanish at lengths of the order of the box length of the simulation.
![[Graphics:Images/index_gr_3.gif]](formula1.gif) (1)
(1)
The fit parameters for Ga and As were taken from [p1].
Since I am interested in stable configurations only, I allowed adatoms to vanish from the surface. As a criterion I took a distance of 2/3 of a unit cell as surface region. Atoms which moved beyond that region are not considered to be surface atoms and thus are left out in the analysis.
The simulations were performed on the
Engenieering Workstations. The code was compiled with the GNU GCC
compiler. I used the built-in random number generator with seeds
taken from [c3].